Peng He

He, Peng
何 鵬
| U of Hong Kong | 1st Honor, BSc in Statistics and Biochemistry | 2009-2012 |
| Caltech | PhD in Genetics | 2012-2019 |
| EMBL-EBI and Sanger | Postdoc | 2019-2024 |
| U of Cambridge | St Edmund's junior research fellow | 2019-2024 |
A more detailed timeline of me is here.
In 2024, I will start my research group at UCSF (Department of Pathology) as an assistant professor.

How is our body built?
Our organs and tissues coordinate various types of cells which move, talk, grow, sleep, mature, and die. What instruct these processes are ultimately programmed in the DNA of the organism. And sitting at the core of these programs are the gene regulatory circuits. To understand these circuits, genomicists have worked very hard searching for correlations between snippets of the codes and biological traits, either natural or artificially induced ones by perturbation.
I am one of them, coming from a quantitative background.
My PhD journey started with the efforts using CRISPR-cas9 proteins to delete a novel type of DNA regions (called lincRNA genes) to watch how cells change their states and molecular signatures. I observed spontaneous differentiation as well as profound stochasticity from clone to clone.
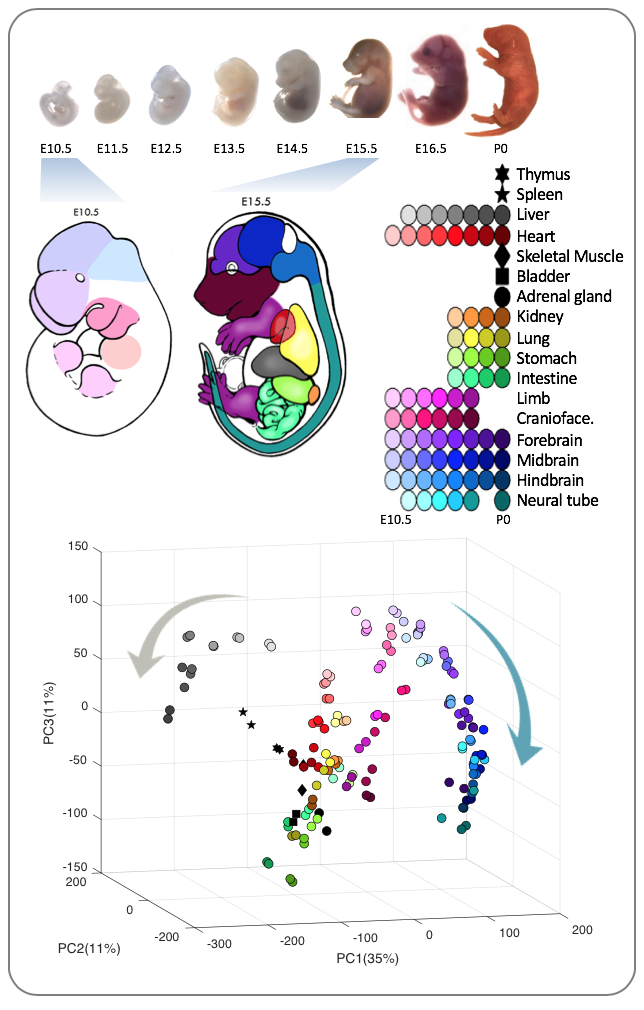
In my second PhD project which became my thesis project, we acquired molecular snapshots of natural developmental processes in the mouse embryo to monitor changes in their gene expression profiles and associated chromatin changes, at both whole-tissue and single-cell levels (as part of the ENCODE project). I used a customized divide-and-conquer scheme to annotate co-expression modules/clusters, followed by data integration cluster-by-cluster and discovered cluster-specific regulatory mechanisms. To further leverage the bulk data, we then built a developmental forelimb atlas of the mouse, and I demonstrated the possibility of using single-cell RNA-seq and bulk chromatin assays to infer cell type-specific cis-regulatory elements and signatures. These results provided a novel framework to study development and gene regulation based on consortium-level bulk and single-cell profiles
As a computational biologist, I independently developed a whole workflow of single-cell transcriptome analysis on MATLAB along with a novel "DeepTree" algorithm to effectively select feature genes based on co-expression. I also collaborated with multiple biochemists and developmental biologists, to study the DNA codes in a variety of physiological and biochemical contexts in mouse, drosophila, human cell lines, C. elegans and sea urchin.
In my postdoctoral study with Dr. Sarah Teichmann and Dr. John Marioni as part of the Human Cell Atlas (HCA) effort, I had the opportunity to extend my PhD work in mice to humans. By integrating human and mouse limb atlases using an in-house workflow I optimised, I identified profound conservation as well as temporal differences in ectoderm maturation and haematopoiesis between mice and humans. I further worked with our experimental team to design complementary spatial transcriptomic experiments to profile the whole-transcriptome at moderate spatial resolutions (10X Visium) and a selected 90-plexed gene panel measured at subcellular resolution (in-situ sequencing). I integrated transcriptome signatures to both types of spatial data using existing and novel ways and dissected finer structures in classical spatial domains such as the distal mesenychme (or “progress zone”). This work not only uncovers novel cell states and spatial regions in limb development, but also provides a key resource for developmental and evolutionary biology communities.
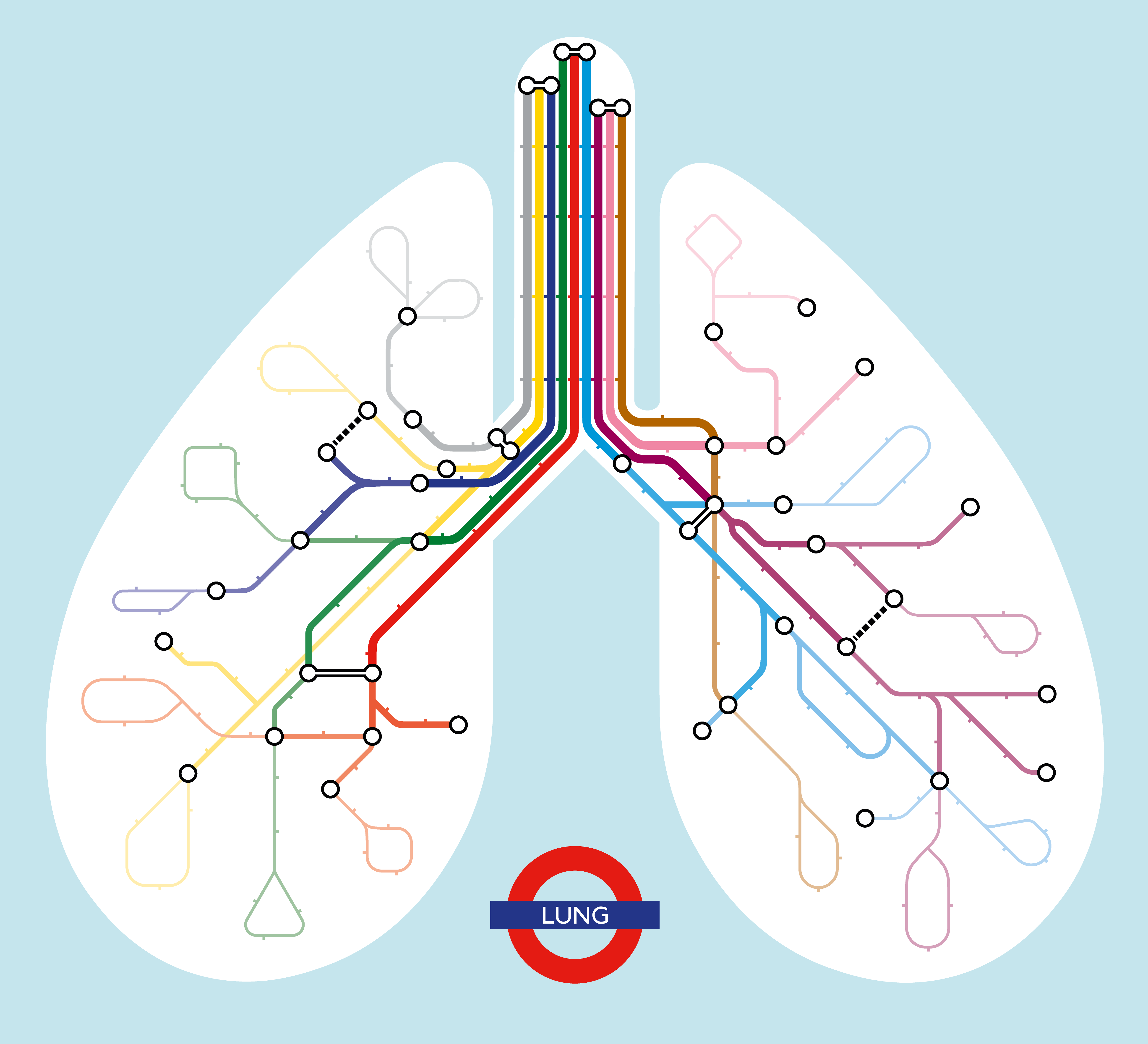
In my second postdoctoral project, our team constructed a comprehensive cell atlas for the human developing lung. I developed a novel approach to aggregate datasets with a soft batch correction and a doublet cluster detection algorithm to complement existing ones, which resulted in a detailed map of 144 cell supported by literature and our in-situ validation experiments. Curiously, in the fine-grained lung atlas I identified multiple novel cell fate paths, especially the ones leading to a novel GHRL+ neuroendocrine subtype first observed by Cao et al. I further matched this new NE subtype to the NEUROD1+ type of small-cell lung cancer (SCLC) cells by transcriptome signature mapping. Furthermore, by integrating scRNA-seq data with scATAC-seq profiles of our matching sample materials, I identified key regulators for this novel cell state which passed our organoid-based functional sufficiency test. This work will provide a community resource for human lung development studies and insights for studying human SCLC.
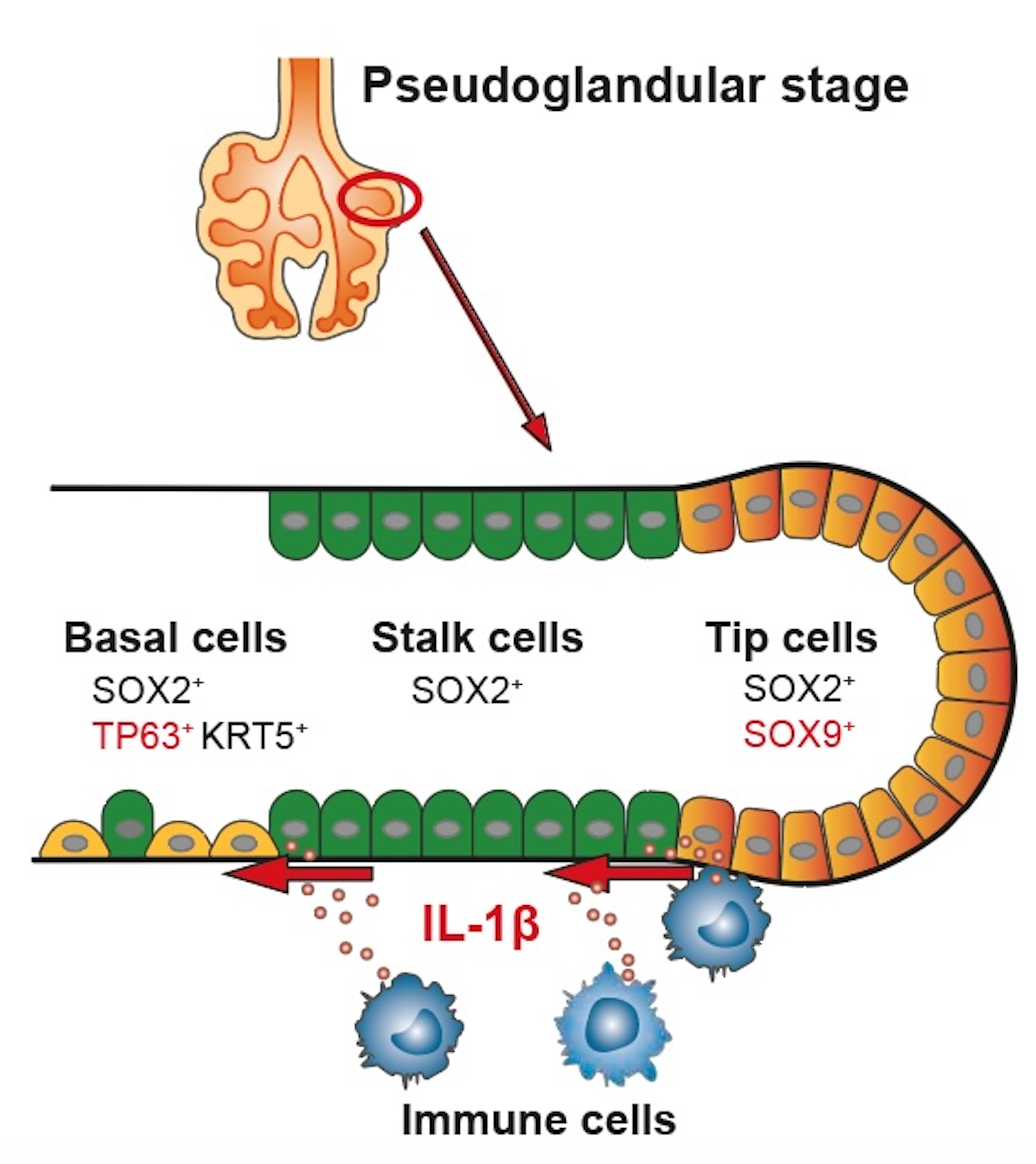
In my third postdoctoral project, our team synthesized a focused immune cell atlas of the human developing lung. We completed the full picture of B cell development in the early lungs and validated the presence of various resident immune cell types. Our collaborators further proved that the cytokines IL-1ß and IL-13 secreted by immune cells promote lung epithelium maturation, underlining the importance of signalling pathways in cell fate decision.
Mathematics in Biology Bi195
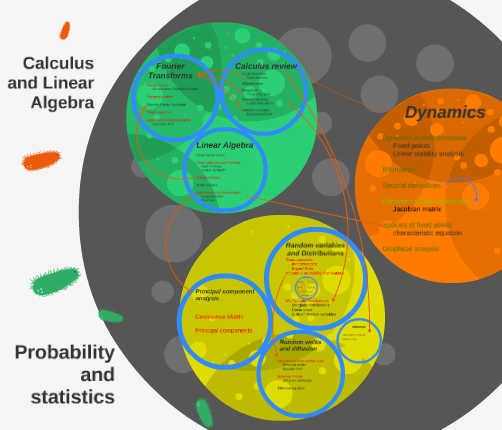
Transplanted from Harvard to Caltech and instructed by Prof. Markus Meister for many years, this course provides an opportunity for biologists to get some basic trainings in linear algebra, calculus, statistics and dynamics which they might use in diversed fields of bio-research.
The image on the left is a snapshot from my TA presentations, showing a rough structure of the course. For more information, please visit our course website and student feedback report
Related courses I have also taught include:
* Introduction to Computational Biology and Bioinformatics Bi181
* Human Genetics and Genomics Bi188
In the past, I had served on committees for Caltech C and Caltech Y Outdoor group. I had also founded Caltech Bioinformatics Club.
Additionally, I have been a newage composer (check out the demo below), pop song chord database manager and analyst, "cactus guy" of community garden, Chinese restaurants and recipes experimentalist, Caltech Glee Club baritone (and webmaster), half-marathon runner (1h 54min), manager of Mathematical Modeling Innovative Practice Base Team which won the 2012 ICM outstanding prize and a Youtuber with more than 2M views.
phe在caltech.edu
ph532于cam.ac.uk
Mount Pleasant Halls, Cambridge, CB3 0BL, UK